As a reader of this blog, you are presumably interested in modelling and are likely to know of at least some benefits of models and modelling in IT and computing. But can you confidently say—and justify!—which modelling language is the best modelling language? And would you support a call for modelling as a specialisation in an IT or computing degree programme, if not deserving to be a separate discipline outright? If so: why? What sets it apart and what are recurring themes across the various types of models and ways of modelling?
They are easy questions to ask, yet non-trivial to answer. I tried anyway. My latest attempt has just been published in the book entitled “The what and how of modelling information and knowledge: from mind maps to ontologies”. The remainder of this post draws from some of its content.
Contents
- Modelling for domain experts: a book on where to start, with what, and how;
- Descriptions and comparisons of five types of models: mind maps, models in biology, conceptual data models, ontologies, and Ontology;
- Deeper reflections for modellers: which one is good for what, why developing an ontology is harder than creating a conceptual data model, ethics and bias in models, and DIY designing your own language.
- References
Modelling for domain experts: a book on where to start, with what, and how;
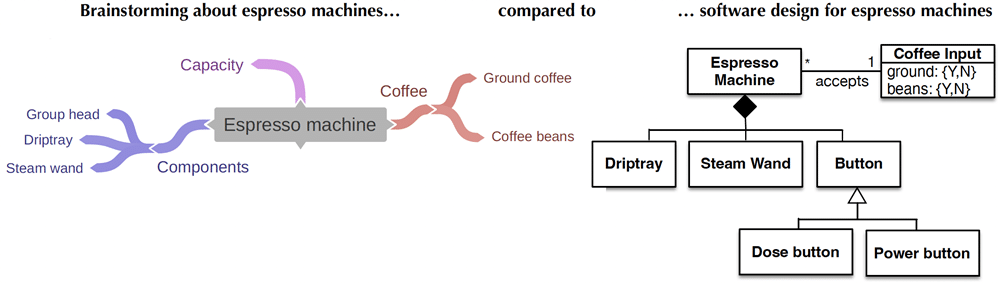
A considerable part of the book is devoted to a journey from where domain expert tend to start modelling—mind maps—to ever more advanced and expressive modelling—all the way up to Ontology (philosophy)—where each successive type of model discussed solves one or more problems of the preceding one. What en passant does happen along the way, is assessing various advantages and limitations, illustrating where each type of model is typically used, and demonstrating recurring principles to designing models. Approaches, methods, and techniques from computing and IT can be transferred to mind maps, biological models, and even to the development of a theory of Ontology.
Descriptions and comparisons of five types of models: mind maps, models in biology, conceptual data models, ontologies, and Ontology;
Strolling along the distinct types of models is also a prelude towards answering the question of which one of them would be the best one. To do so, I take two ways of evaluation: a feature-based comparison and a task-based comparison.
A feature-based comparison
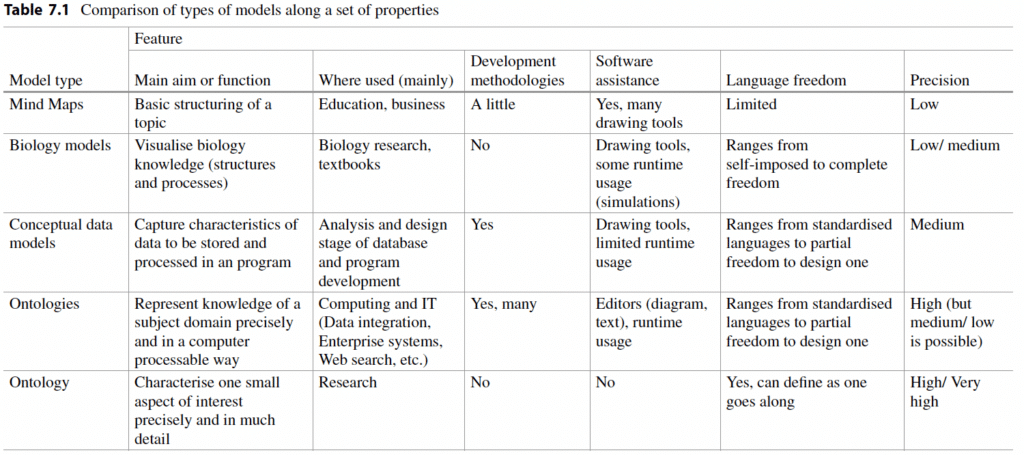
For the feature-based comparison, one can of course debate about which features to select. Here, I keep it short by just mentioning them: which aim each type of model serves, where they are typically used, to what extent is there methodological and software support, how extensible the modelling languages are, and what level of precision can be obtained. The outcome of the comparison is shown in the table below and discussed in more detail in the book.
Perhaps unsurprisingly, the comparison shows that there is no single best type of model for all aims in the general case. What it may aid in, is a professional judgement when to use which one. For instance, it’s fine to develop an ontology to integrate electronic health record data for research into symptoms and treatments of a disease [Reese23] or generating ontology-based specialist educational questions [RaboanaryEtal22]. But it would be irresponsible to push for ontology development if brainstorming for ideas to solve an urgent problem is all that is asked for; then mind maps will do just fine. If you need to design a database, Ontology won’t get you there, but a conceptual data model can, especially when there is some tolerance for imprecision. And so on and so forth.Notwithstanding that they are distinct types of models with their own purposes, cross-fertilization to support each other is very well possible: to create a conceptual data model for an ontological theory as part of a procedure to develop either an ontology or do an ontological investigation, and conceptual modelling notations are (ab)used for sketching an ontology’s content. Conversely, outcomes from Ontology (philosophy) inform the practice of, and languages used for, conceptual modelling. And many a biological model can be recast as a domain-specific language. Mind maps may not have as many usages beyond itself, but mind mapping can, and has been, used in the initial stages of ontology development.
A task-based comparison: learning about labour migration
The task-based comparison revisits the educationalists’ mantra that mind maps are great. The reader easily can repeat the task. As use case for the task of learning a page’s worth of content, I used a textbook page on labour migration to (try to) create a model for it. Which type of model fares best to complete that task?
In this instance, it is possible to provide evidence for the best one for the task, and the winner turns out to be the conceptual data model. Why? The mind map is superficial and imprecise, where nuances required for that level of learning slipped through. Biological modelling doesn’t work well with the abstract content of labour migration. And developing an ontology requires a modeller to go way beyond the page’s content. The latter is good in other learning contexts where critical analysis is asked for [Bertens22], higher up in Bloom’s taxonomy and in more advanced classes in a degree programme, but that was not the task at hand. Nevermind the ontology of concepts such as labour and foreigner. The conceptual data model did have the right mix of permitting some detail and precision yet not needing to go beyond the content.
Deeper reflections for modellers: which one is good for what, why developing an ontology is harder than creating a conceptual data model, ethics and bias in models, and DIY designing your own language.
All this does support the idea that there may well be enough commonalities to at least to set up modelling as a specialisation of a degree programme, with evidence. Jordi Cabot and Antonio Vallecillo propose to go a step further, to have modelling as a separate discipline [CabotVallecillo22]. I think we’re not there yet, but this book, written independently in parallel to their efforts, does provide ample evidence that we’re moving in that direction. The modelling that we do in IT and computing, and how we do it, is largely transferrable to modelling in biology and to mind mapping, and it even can help structuring an ontological investigation. DSLs also appear to have a bright future for especially the myriad of biological models, so that we can provide tooling support. This can facilitate analysing the models automatically instead of the manual labour we did [KeetFillottrani15] or having to set up a laborious OCR, ML and NLP pipeline by others [HanspersEtAl21]. That, in turn, may foster more new insights facilitated by modelling and inform new research avenues on modelling. And so, the work continues.
p.s.: The book is available from Springer, Springer professional, many national and international online retailers selling the softcover hardcopy and the eBook (e.g., Amazon), as well as university libraries.
References
[Bertens22] Bertens LMF. Modeling the art historical canon. Arts Humanit Higher Educ, 2022, 21(3):240–262.
[CabotVallecillo22] Cabot J, Vallecillo A. Modeling should be an independent scientific discipline. Softw Syst Model, 2022, 22:2101–2107
[HanspersEtal21] Hanspers K, Riutta A, Summer-Kutmon M, Pico AR. Pathway information extracted from 25 years of pathway figures. Genome Biol, 2021, 21:273
[KeetFillottrani15] Keet, C.M., Fillottrani, P.R. An analysis and characterisation of publicly available conceptual models. 34th International Conference on Conceptual Modeling (ER’15). Johannesson, P., Lee, M.L. Liddle, S.W., Opdahl, A.L., Pastor Lopez, O. (Eds.). Springer LNCS vol 9381, 585-593. 19-22 Oct, Stockholm, Sweden
[RoboanaryEtAl22] Raboanary, T., Wang, S., Keet, C.M. Generating Answerable Questions from Ontologies for Educational Exercises. 15th Metadata and Semantics Research Conference (MTSR’21). Garoufallou, E., Ovalle-Perandones, M-A., Vlachidis, A (Eds.). 2022, Springer CCIS vol. 1537, 28-40.
[Reese23] Reese, J. et al. Generalisable long COVID subtypes: findings from the NIH N3C and RECOVER programmes. eBioMedicine, Volume 87, 104413, January 2023.



Recent Comments