In this post, written in collaboration with Yoram Reich, I discuss how the analysis of process descriptions can improve the quality of research practices.
Process descriptions are essential to modern life. When we try to assemble a piece of furniture, prepare a culinary dish, develop a product or execute a scientific experiment, we typically look for process descriptions to guide us. Descriptions may be in form of installation instructions, dish recipe, project development plans or experiment protocols.
Research papers typically tell the story of a conducted research, and, as such, they are a form of process descriptions. The scientific research community is seemingly obsessed with the quality of its reports. Scientific research process quality is meant to assure core scientific values, and particularly reproducibility and theory building. Alas, the long-established peer-review process is primarily concerned with the end results and their novelty, with the quality of the scientific process report not being rigorously validated.
Multiple efforts are made by the scientific community to improve the quality of research reports. These efforts typically yield recommendations in the form of guidelines and checklists. A few examples can be found in the works of the TOP (Transparency and Openness Promotion committee) [1] and of the EQUATOR (Enhancing the QUAlity and Transparency Of health Research) international initiative [2]. Various publications adopt such recommendations (see, for example,[3]). Still, natural language guidelines and checklists are insufficient for assuring report quality. Replication efforts are considered the ultimate way to examine reproducibility, yet these are demanding with respect to resources and traditionally not well rewarded in scientific culture. The scientific community keeps trying to solve its quality related issues with its own tools, “inside its box,” while potential solutions may lie outside it.
Model-based analysis using PROVE
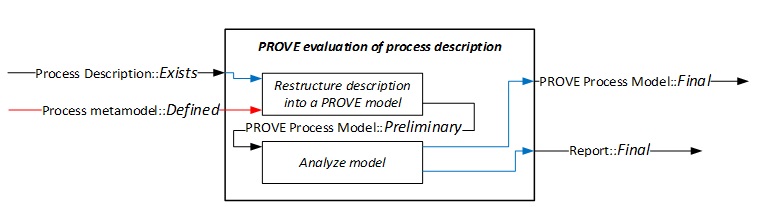
In our research – recently reported in IEEE Systems Journal [4] – we suggest a different take on solving the problem, using a systems engineering perspective. Identifying that scientific research is in fact a man-made (engineered) system, we claim it can be improved by harnessing the engineering body of knowledge. Specifically, we tried to apply elements of our modeling framework PROVE (Process Oriented Viewpoint for Engineering) – originally developed for designing development processes [5] – to published research-related process descriptions. PROVE is a domain-specific modeling framework, including dedicated graphical notation that is aligned with the development process ontology and concepts. We consider research processes as development processes, as they primarily aim to develop new scientific knowledge (this can be theoretical knowledge, methods, tools, etc.). Accordingly, we apply PROVE to restructure free-form process descriptions into a formal model, and analyze the restructured descriptions based on desired process ontology and model paradigms.
Demonstration
In the IEEE System Journal paper, we report representative findings from two analyses and demonstrate how these can be improved to promote their reproducibility and, consequentially, scientific advancement. For the readers of Modeling Languages, we briefly discuss two different, unpublished analyses of research related process descriptions taken from leading scientific journals. These are simple examples that further demonstrate the problematic report of processes in research publications, and that the impact of these problems – such as those shortly mentioned – is intensified in large scale, complex and/or collaborative research. These analyses were conducted using PROVE Tool, which is an Eclipse-based modeling tool that implements PROVE and is available for free use (and is designated to be released as an open-source tool by the end of this year) [6].
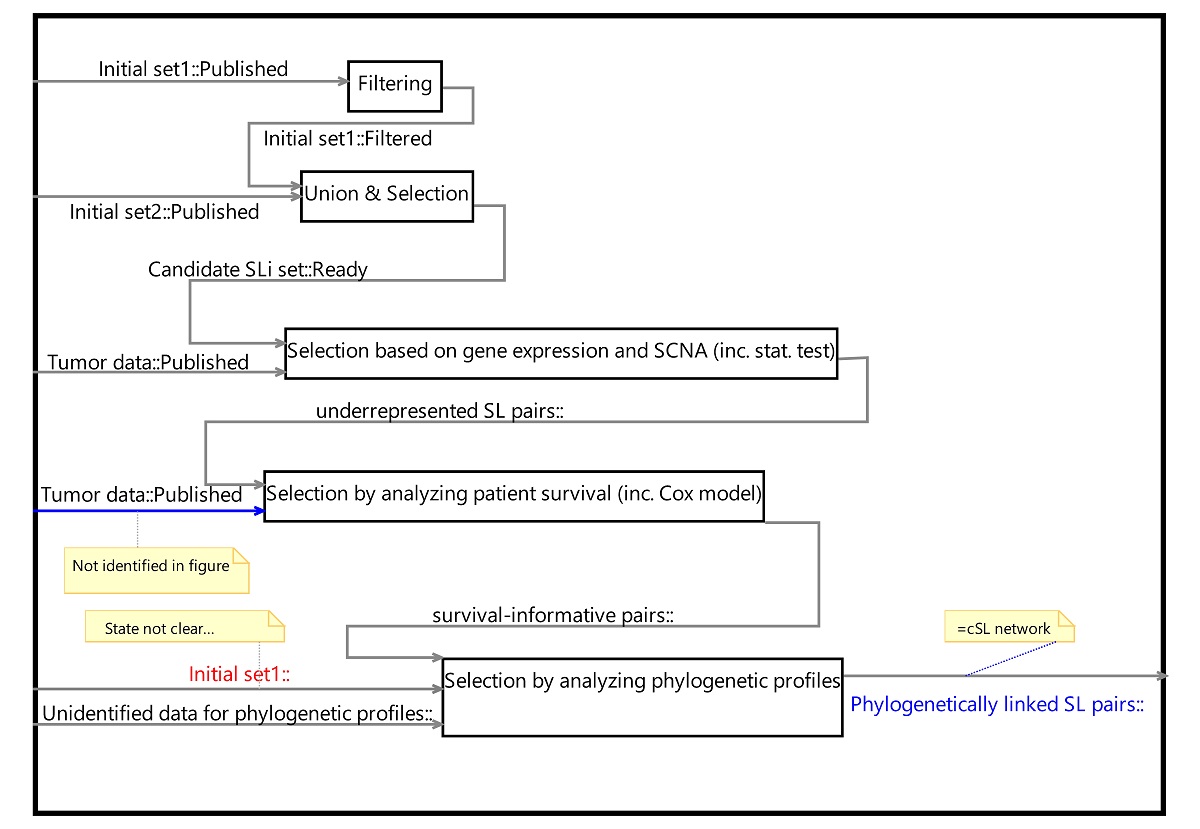
The first analysis is of a computational biology paper published in Nature Communications in 2018 [7]. Figure 1 of the paper and the pertinent description in the text were restructured as a PROVE model, and it is reproduced here. Three notes (in the figure below) mark our main findings:
- Based on the text, published tumor data is required for the “selection by analyzing patient survival…” but this dependency is not captured in the free-form figure. This indicates an inconsistency between the natural language description and the figure content.
- The activity “Selection by analyzing phylogenetic profiles” requires an initial set of data denoted “Initial set1”; yet the process description does not specify the specific set. Had the set appeared only once in our process description, this would have been trivial to understand; however, the set appears twice!: once as “Published” and once as “Filtered.” Obviously, computations applied to different sets of data yield different results.
- There is terminology inconsistency between the figure, which terms the final process product as “cSL network,” and the text, which uses the term “Phylogenetically linked SL pairs” for the product.
These findings show that the process mentioned in the analyzed paper is somewhat ambiguous. Accordingly, an attempt to replicate the process is likely to require additional knowledge as well as support from the original authors – an issue highlighted by many replication studies.
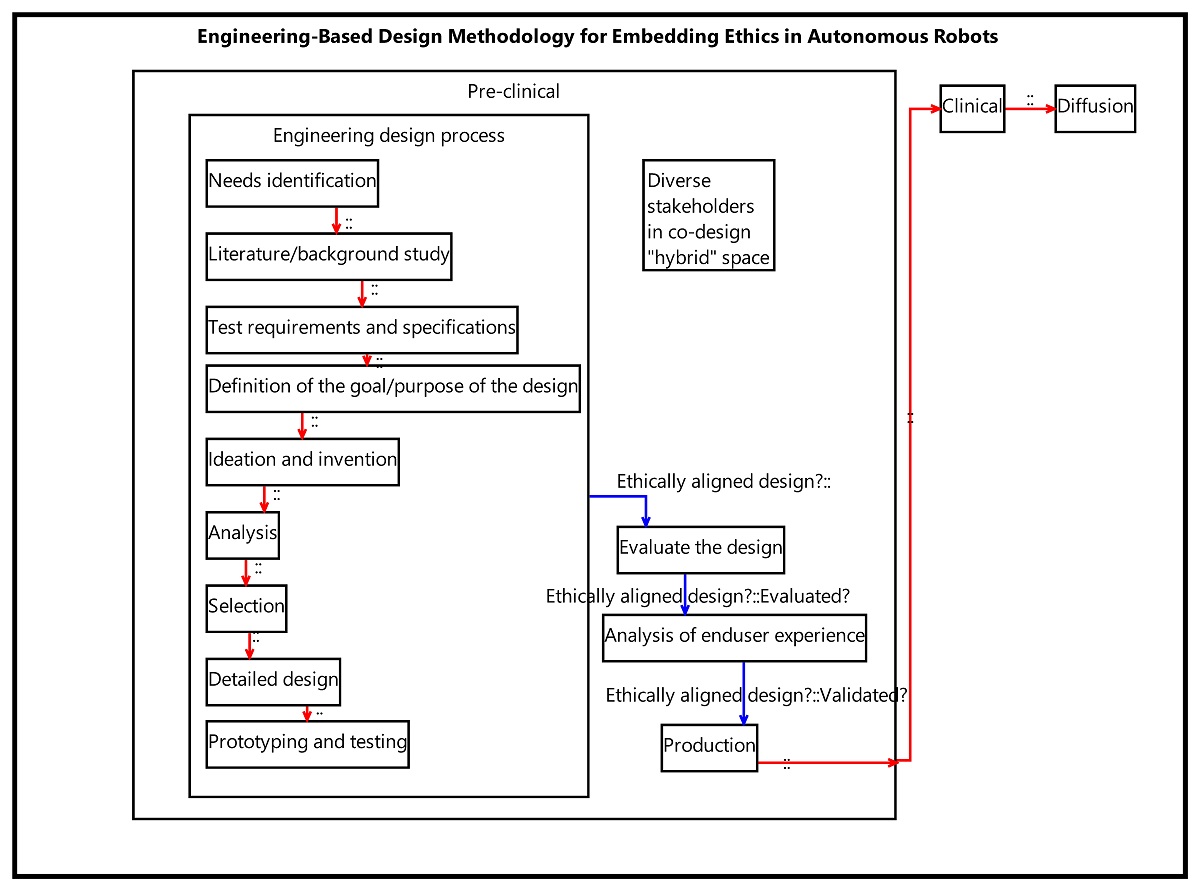
The second analysis is of a suggested methodology for embedding ethics in autonomous robots, published in Proceedings of the IEEE in 2019 [8]. The proposed methodology – captured in Figure 2 of the paper and its corresponding text – was restructured as a PROVE model, shown below. Our model indicates that the proposed methodology – as described in the paper – is insufficient as a protocol for practical use. This is particularly evident in the blank, unidentified artifact flows between the various activities. This also suggests that there is a lot more room for future improvement, which is, in fact, an opportunity for future research! This demonstrates how PROVE analyses may promote theory building. In comparison, our analysis of the C-K design theory process description, which was reported in the aforementioned IEEE Systems paper, found the description to be disciplined and rigorous and is, therefore, more likely to be applicable. Nevertheless, by using domain-specific model-based analysis, we managed to propose a specific improvement to the theory (only to find out it has already been incorporated into the theory, after few years of additional research), as reported in the supplementary material of our paper [4].
Summary
In conclusion, we believe that the scientific community should embrace engineering knowledge and tools to systematically improve its performance and design. We invite researchers to discuss this with us and welcome invitations to collaborate and make this happen!
References
- A. Nosek, G. Alter, G. C. Banks, D. Borsboom, S. D. Bowman, S. J. Breckler, S. Buck, C. D. Chambers, G. Chin, G. Christensen, M. Contestabile, A. Dafoe, E. Eich, J. Freese, R. Glennerster, D. Goroff, D. P. Green, B. Hesse, M. Humphreys, J. Ishiyama, D. Karlan, A. Kraut, A. Lupia, P. Mabry, T. Madon, N. Malhotra, E. Mayo-Wilson, M. Mcnutt, E. Miguel, E. Levy Paluck, U. Simonsohn, C. Soderberg, B. A. Spellman, J. Turitto, G. Vandenbos, S. Vazire, E. J. Wagenmakers, R. Wilson, T. Yarkoni, “Promoting an open research culture”, Science 348, 1422–1425, 2015.
- EQUATOR (Enhancing the QUAlity and Transparency Of health Research) international initiative website: https://www.equator-network.org/
- Montenegro‐Montero, A. L. García‐Basteiro, “Transparency and reproducibility: A step forward.” Health science Reports 2, no. 3, 2019.
- Shaked, Y. Reich. “Improving Process Descriptions in Research by Model-Based Analysis”, IEEE Systems Journal, 2020. https://doi.org/10.1109/JSYST.2020.2990488
- Shaked, Y. Reich. “Designing development processes related to system of systems using a modeling framework”, Systems Engineering, 2019. https://doi.org/10.1002/sys.21512
- PROVE Tool at the TAU-SERI github repository: https://github.com/TAU-SERI/PROVE
- Lee JS, Das A, Jerby-Arnon L, Arafeh R, Auslander N, Davidson M, McGarry L, James D, Amzallag A, Park SG, Cheng K. “Harnessing synthetic lethality to predict the response to cancer treatment”, Nature communications, 2018. https://doi.org/10.1038/s41467-018-04647-1
- Robertson LJ, Abbas R, Alici G, Munoz A, Michael K. “Engineering-based design methodology for embedding ethics in autonomous robots”, Proceedings of the IEEE, 2019. https://doi.org/10.1109/JPROC.2018.2889678

An enthusiastic systems engineering researcher and practitioner, with a PhD from Tel Aviv University and extensive experience in industry.



Recent Comments